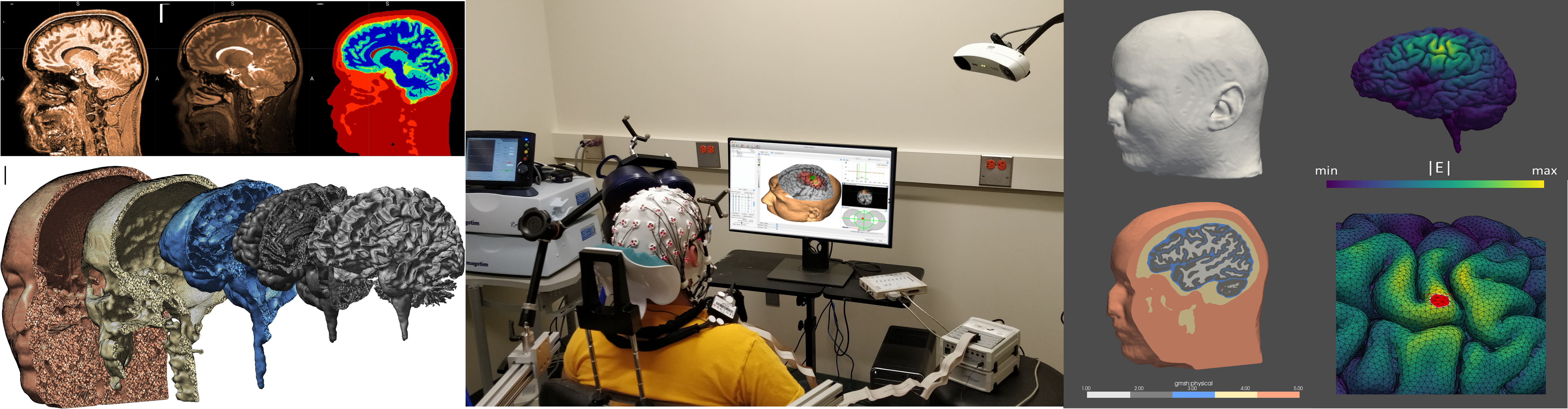
Electric Field Simulation Software
SimNIBS is a user-friendly modeling tool for calculating the electric field induced in the brain by non-invasive brain stimulation. Additional features, and improved visualization and graphical user interface are currently being developed for SimNIBS.
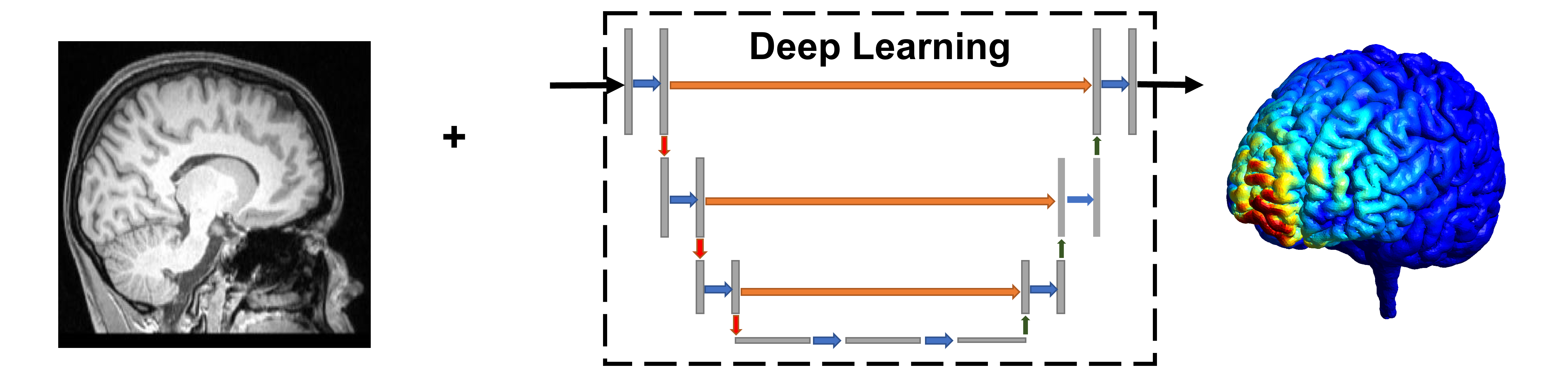
Deep-Learning Based TMS Modeling
Using current state-of-the-art numerical methods, we are able to develop computational models depicting how Transcranial Magnetic Stimulation (TMS) will induce an electric field in a unique head model. While this method does produce highly accurate results, the process is time-consuming which has resulted in a bottleneck in research and limited clinical feasibility. A deep-learning-based method can generate computational models rapidly to allow for more adaptive research and clinical protocols.
Brain Stimulation for Stroke Recovery in Children
Ischemic perinatal stroke affects as many as 1 in 2,300 live births. TMS and transcranial electric stimulation (TES) have shown promise as noninvasive cortical assessment and neuromodulation techniques for stroke rehabilitation. We are integrating individual realistic head models into the therapy to optimize neuromodulation targets for rehabilitation in perinatal stroke. Using our novel computational method, we are able to develop more precise and effective personalized treatments for stroke rehabilitation.
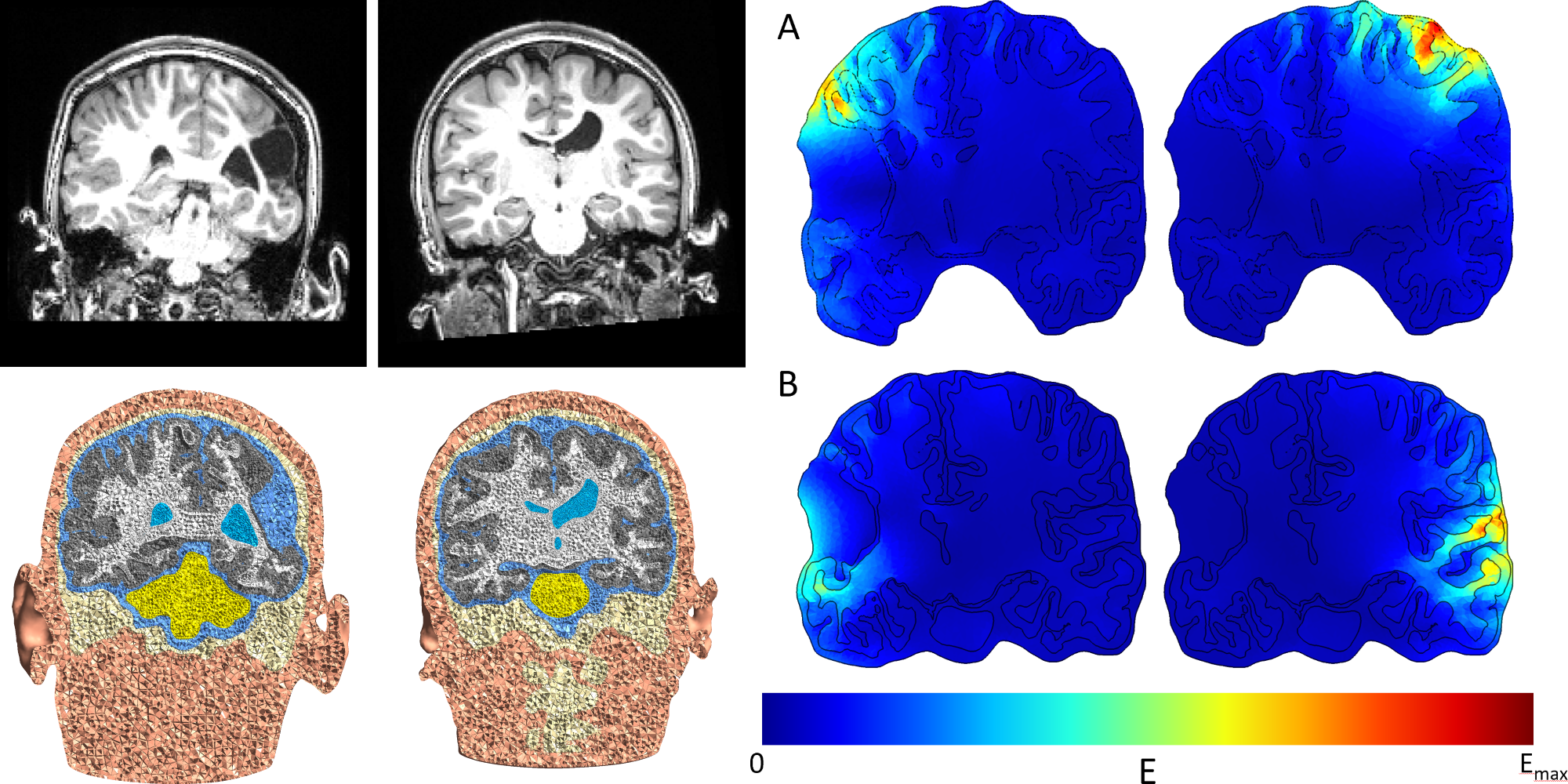
(Left) Individual head models from patients' MRI are made using the finite element method (FEM). Anatomically accurate representation of a lesion is a crucial step for patient-specific modeling. (Right) Simulation of the electric field during TMS over the lesioned (left) and non-lesioned (right) hemispheres. A) TMS coil over the motor cortex. B) TMS coil over the temporal lobe. We conduct FEM simulations to understand how electric fields change for different brain anatomy and in the presence of a lesion.
Computational Modeling of Neural Responses to tACS in a Neuronal Network Model
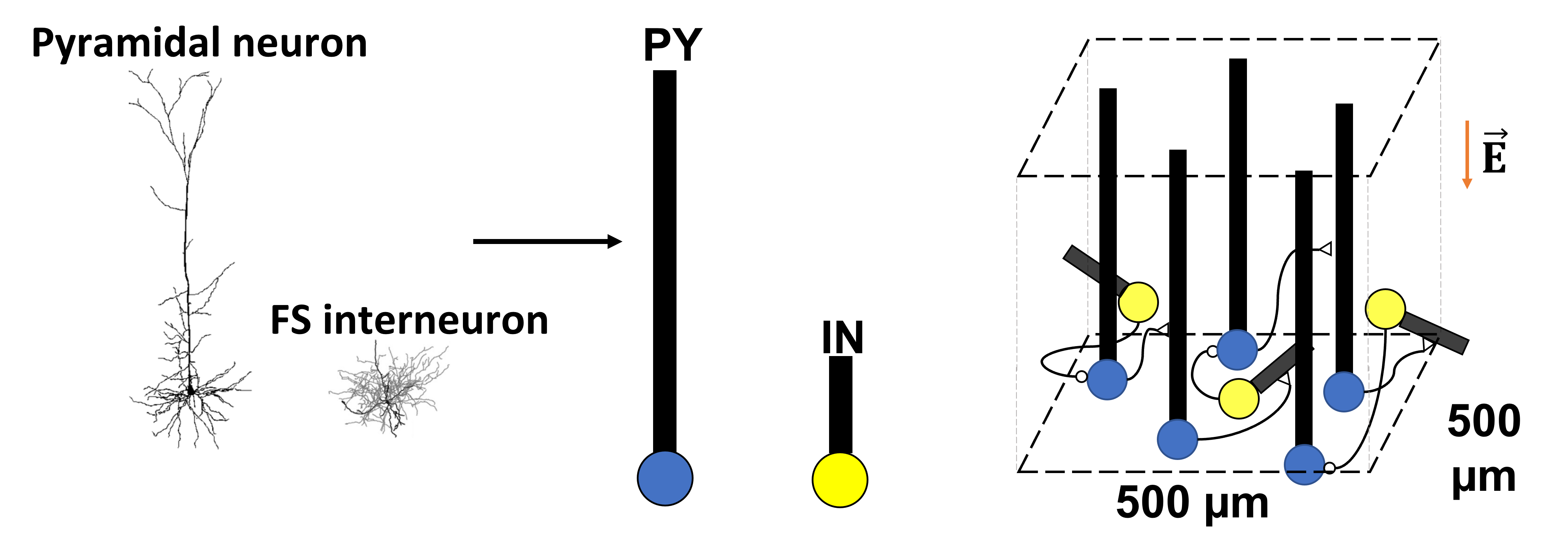
We are developing a neuronal network model which consists of 1000 two-compartment neurons (800 excitatory pyramidal neurons and 200 inhibitory interneurons). These neurons are spatially arranged and connected to match the realistic neuron distribution in cortical layers. Then, we explore different parameters in this neuronal network to probe how single neurons and neuronal populations respond to the electric field.
Multi-Scale Modeling from Whole Brain to Single Neuron Level
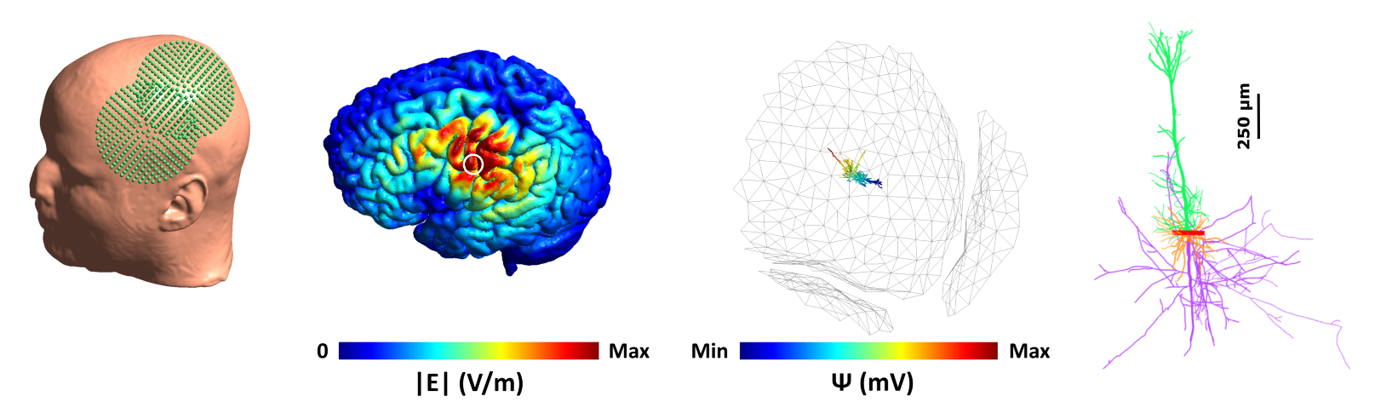
Multi-scale modeling can complement experimental research by providing a framework between the physical input parameters of TMS and the cellular and subcellular neural effects of TMS. We developed a multi-scale Neuron Modeling for TMS toolbox (NeMo-TMS) that enables researchers to easily generate accurate neuron models from morphological reconstructions, couple them to the external electric fields induced by TMS, and to simulate the cellular and subcellular responses of the neurons.
Understanding the effects of electric stimulation on neural activity of single cells
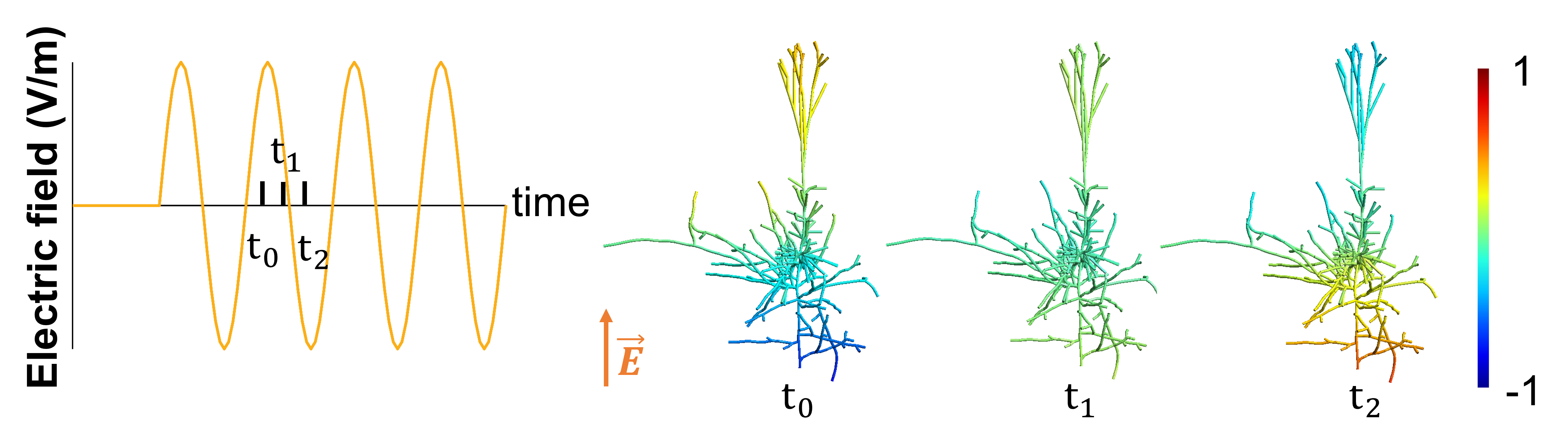
We seek to understand how different types of electric stimulation (Transcranial Alternating Current Stimulation (tACS), Transcranial Direction Current Stimulation (tDCS), or TMS) affect the neural activity of single neurons (neural entrainment or firing pattern for example). We investigate the effects of tACS on neural entrainment in single neocortical neurons exhibiting complex morphologies. These models include ongoing firing activity as well as realistic ionic channel distribution. We also perform in-vivo recordings of single units in awake non-human primates while applying electric stimulation.
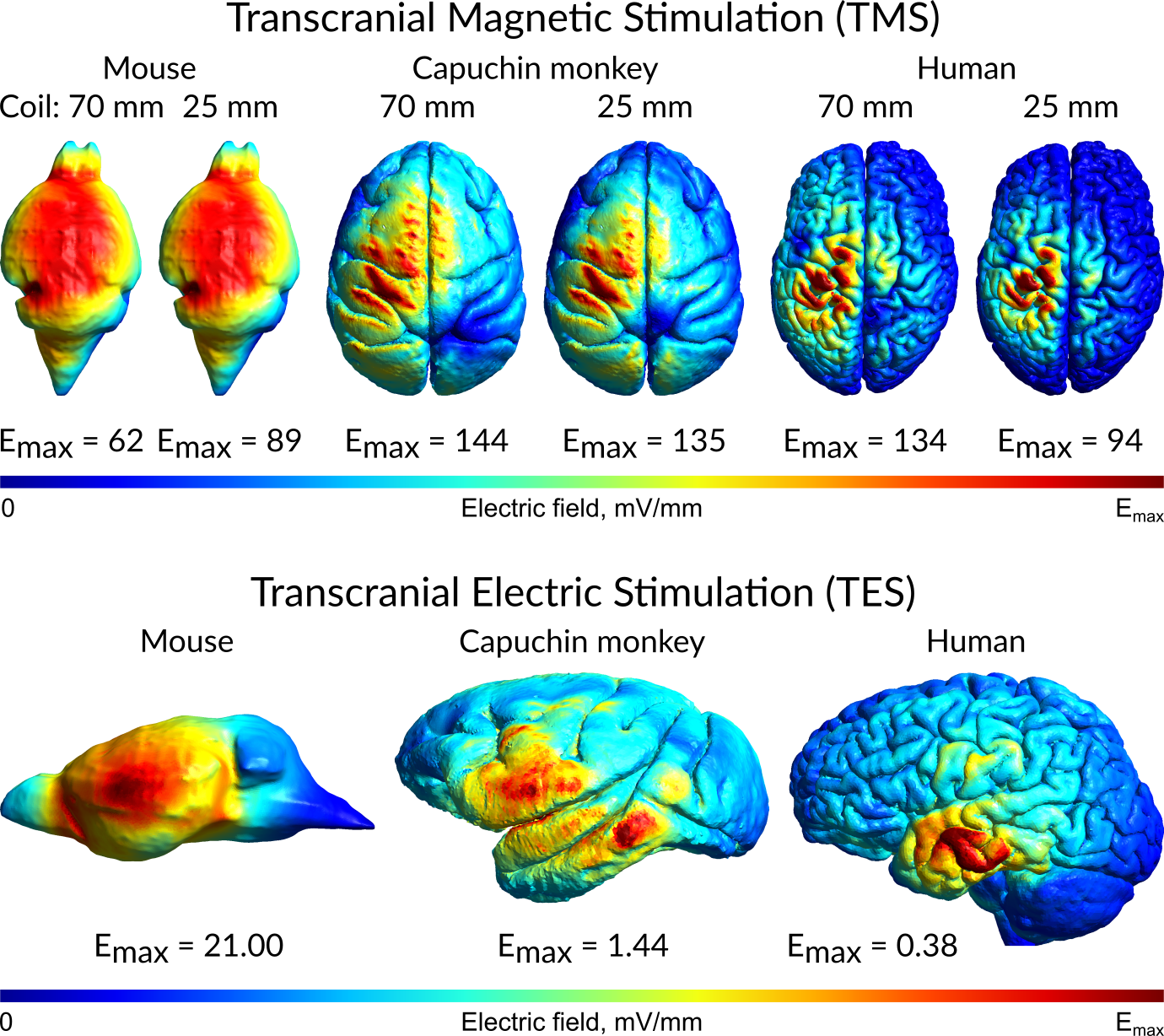
Cross-Species Modeling for Translation Research
Our lab builds nonstandard 3D models to run Non-Invasive Brain Stimulation (NIBS) simulations. We study things like how stroke lesions, different size heads, the inclusion of additional tissue types, etc. affect NIBS-generated electric fields. Our goals are to better understand dosing and precision targeting and to build more accurate models.